3437 Mary Sue Coleman Hall, 210 Washtenaw Ave
Ann Arbor, MI 48109
Available to mentor

Janet Smith's research focuses on understanding biological processes through knowledge of the structures of key protein molecules. Early in her independent career, she made major contributions to the understanding of catalysis and regulation in glutamine amidotransferases, phosphoribosyltransferases and photosynthetic proteins by solving and interpreting crystal structures of several proteins of each type. She has also contributed to the development of methods for rapid determination of protein crystal structures, particularly using synchrotron X-ray sources.
A native of Pennsylvania, Smith studied chemistry as a National Merit Scholar at Indiana University of Pennsylvania. Finding biochemistry to be the most stimulating area of chemistry, she continued her study in that field at the University of Wisconsin-Madison where she was convinced of the importance of structure in biology during her research with advisor M. Sundaralingam. Smith then pursued a growing interest in protein structure as a postdoc with Wayne Hendrickson as a National Research Council Research Fellow at the Naval Research Laboratory and as associate research scientist at the Howard Hughes Medical Institute at Columbia University.
Smith established an independent research program in structural biology at Purdue, where she remained as a professor of biological sciences until moving to the University of Michigan and the LSI. She has been a visiting scientist at the European Molecular Biology Laboratory and the European Synchrotron Radiation Facility in Grenoble, France, and a lecturer at numerous international schools on structural biology and synchrotron radiation. She is also a frequent advisor to synchrotron radiation facilities and synchrotron structural biology labs both in the U.S. and abroad.
Smith Lab
-
Center MemberRogel Cancer Center
Our group studies protein structure in order to understand the molecular mechanisms of biological processes and to develop testable hypotheses about function. The lab currently has two research focuses.
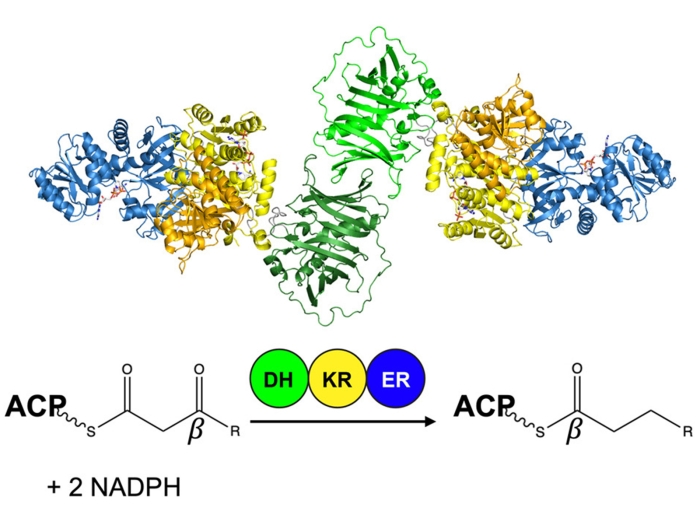
We study the enzymes of natural product biosynthesis with a focus on modular polyketide synthases. One focus of our research is the mechanism of throughput and the substrate/product selecitvity of assembly-line megasynthases. Another is the adaptation of “ordinary” enzymes of primary metabolism to new chemical transformations that enrich nature’s chemical toolbox and have potential as biocatalysts.
Our studies of the pathogenesis of RNA viruses include both viral and host proteins. Of the host proteins, the zinc-finger antiviral protein recognizes viral RNA in the cytoplasm and targets it for destruction by host nucleases. Other proteins restrict retroviruses by hypermutation of the viral DNA product of reverse transcription. A rapidly evolving viral protein targets the restriction factors for destruction by the proteasome. A multi-functional flavivirus protein helps the virus escape detection by the immune system, increases the infectivity of virus particles, and also has an essential role in viral RNA replication.
-
Mydy LS, Hungerford J, Chigumba DN, Konwerski JR, Jantzi SC, Wang D, Smith JL, Kersten RD. Nat Chem Biol, 2024 Feb 14;Journal ArticleAn intramolecular macrocyclase in plant ribosomal peptide biosynthesis.
DOI:10.1038/s41589-024-01552-1 PMID: 38355722 -
McCullough TM, Dhar A, Akey DL, Konwerski JR, Sherman DH, Smith JL. Structure, 2023 Sep 7; 31 (9): 1109 - 1120.e3.Journal ArticleStructure of a modular polyketide synthase reducing region.
DOI:10.1016/j.str.2023.05.019 PMID: 37348494 -
Chiang C-H, Wymore T, Rodríguez Benítez A, Hussain A, Smith JL, Brooks CL, Narayan ARH. Proc Natl Acad Sci U S A, 2023 Apr 11; 120 (15): e2218248120Journal ArticleDeciphering the evolution of flavin-dependent monooxygenase stereoselectivity using ancestral sequence reconstruction.
DOI:10.1073/pnas.2218248120 PMID: 37014851 -
Pham VV, Gao M, Meagher JL, Smith JL, D'Souza VM. Commun Biol, 2022 Aug 15; 5 (1): 819Journal ArticleA structure-based mechanism for displacement of the HEXIM adapter from 7SK small nuclear RNA.
DOI:10.1038/s42003-022-03734-w PMID: 35970937 -
Kelly SP, Shende VV, Flynn AR, Dan Q, Ye Y, Smith JL, Tsukamoto S, Sigman MS, Sherman DH. J Am Chem Soc, 2022 Oct 26; 144 (42): 19326 - 19336.Journal ArticleData Science-Driven Analysis of Substrate-Permissive Diketopiperazine Reverse Prenyltransferase NotF: Applications in Protein Engineering and Cascade Biocatalytic Synthesis of (-)-Eurotiumin A.
DOI:10.1021/jacs.2c06631 PMID: 36223664 -
Lao Y, Skiba MA, Chun SW, Narayan ARH, Smith JL. ACS Chem Biol, 2022 Aug 19; 17 (8): 2088 - 2098.Journal ArticleStructural Basis for Control of Methylation Extent in Polyketide Synthase Metal-Dependent C-Methyltransferases.
DOI:10.1021/acschembio.2c00085 PMID: 35594521 -
Stepanov S, Kissick D, Makarov O, Hilgart M, Becker M, Venugopalan N, Xu S, Smith JL, Fischetti RF. J Synchrotron Radiat, 2022 Mar 1; 29 (Pt 2): 393 - 399.Journal ArticleFast automated energy changes at synchrotron radiation beamlines equipped with transfocator or focusing mirrors.
DOI:10.1107/S1600577522001084 PMID: 35254302 -
Wessel AW, Dowd KA, Biering SB, Zhang P, Edeling MA, Nelson CA, Funk KE, DeMaso CR, Klein RS, Smith JL, Cao TM, Kuhn RJ, Fremont DH, Harris E, Pierson TC, Diamond MS. J Virol, 2021 Sep 27; 95 (20): e0084421Journal ArticleLevels of Circulating NS1 Impact West Nile Virus Spread to the Brain.
DOI:10.1128/JVI.00844-21 PMID: 34346770